You're reading an old version of this documentation. For up-to-date information, please have a look at v0.12.
Plot annotations: FillBetween
Classes used:
Models:
Processing:
Plotting:
Annotations:
Graphical representation of data and results is one of the most important aspects of presenting scientific results. A good figure is a figure allowing the reader to immediately catch the important aspects, not relying on reading the (nevertheless always important) caption with more description.
To this end, there is the frequent need to annotate figures, i.e. add additional lines, areas, or even text. This is what can be done with the concrete subclasses of aspecd.annotation.PlotAnnotation.
Here, we focus on filling areas below or between curves that can be used, e.g., to highlight individual components. Suppose you have fitted a spectral line with a series of Lorentzians, and you would like to show the individual components as shaded areas. That’s exactly what aspecd.annotation.FillBetween is designed for.
Recipe
Shown below is the entire recipe. As this is quite lengthy, separate parts will be detailed below in the “Results” section.
1format:
2 type: ASpecD recipe
3 version: '0.2'
4
5settings:
6 autosave_plots: False
7
8tasks:
9 - kind: model
10 type: Zeros
11 properties:
12 parameters:
13 shape: 1001
14 range: [0, 20]
15 result: dummy
16
17 - kind: model
18 type: Lorentzian
19 from_dataset: dummy
20 properties:
21 parameters:
22 position: 5
23 width: 0.8
24 result: lorentzian1
25
26 - kind: model
27 type: Lorentzian
28 from_dataset: dummy
29 properties:
30 parameters:
31 position: 8
32 width: 2
33 result: lorentzian2
34
35 - kind: model
36 type: CompositeModel
37 from_dataset: dummy
38 properties:
39 models:
40 - Lorentzian
41 - Lorentzian
42 parameters:
43 - position: 5
44 width: 0.8
45 - position: 8
46 width: 2
47 result: model_data
48
49 - kind: multiplot
50 type: MultiPlotter1D
51 properties:
52 properties:
53 axes:
54 xlabel: "$position$ / a.u."
55 xlim: [0, 20]
56 ylim: [0, 1.45]
57 drawings:
58 - linewidth: 1.5
59 color: black
60 - linewidth: 1
61 color: gray
62 - linewidth: 1
63 color: gray
64 parameters:
65 tight_layout: True
66 filename: plotting-annotation-fillbetween-no-annotation.pdf
67 apply_to:
68 - model_data
69 - lorentzian1
70 - lorentzian2
71 comment: >
72 Plotter showing just the model.
73
74 - kind: multiplot
75 type: MultiPlotter1D
76 properties:
77 properties:
78 axes:
79 xlabel: "$position$ / a.u."
80 xlim: [0, 20]
81 ylim: [0, 1.45]
82 parameters:
83 tight_layout: True
84 filename: plotting-annotation-fillbetween.pdf
85 apply_to:
86 - model_data
87 - lorentzian1
88 - lorentzian2
89 result: plot-with-fillbetween
90 comment: >
91 Plotter showing the model and the two coloured surfaces.
92
93 - kind: plotannotation
94 type: FillBetween
95 properties:
96 parameters:
97 data:
98 - lorentzian1
99 - lorentzian2
100 plotter: plot-with-fillbetween
101 comment: >
102 Coloured surfaces of the Lorentzians with default properties.
103
104 - kind: multiplot
105 type: MultiPlotter1D
106 properties:
107 properties:
108 axes:
109 xlabel: "$position$ / a.u."
110 xlim: [0, 20]
111 ylim: [0, 1.45]
112 parameters:
113 tight_layout: True
114 filename: plotting-annotation-fillbetween-separate.pdf
115 apply_to:
116 - model_data
117 - lorentzian1
118 - lorentzian2
119 result: plot-with-fillbetween-separate
120 comment: >
121 Plotter showing the model and the two coloured surfaces separately styled.
122
123 - kind: plotannotation
124 type: FillBetween
125 properties:
126 parameters:
127 data:
128 - lorentzian1
129 properties:
130 facecolor: orange
131 alpha: 0.3
132 plotter: plot-with-fillbetween-separate
133 result: fillbetween-lorentzian1
134 comment: >
135 Coloured surfaces of the first Lorentzian.
136
137 - kind: plotannotation
138 type: FillBetween
139 properties:
140 parameters:
141 data:
142 - lorentzian2
143 properties:
144 facecolor: green
145 alpha: 0.3
146 plotter: plot-with-fillbetween-separate
147 result: fillbetween-lorentzian2
148 comment: >
149 Coloured surfaces of the second Lorentzian.
150
151 - kind: singleplot
152 type: SinglePlotter1D
153 properties:
154 properties:
155 axes:
156 xlabel: "$position$ / a.u."
157 xlim: [0, 20]
158 ylim: [0, 1.45]
159 parameters:
160 tight_layout: True
161 filename: plotting-annotation-fillbetween-singleplot.pdf
162 apply_to:
163 - model_data
164 annotations:
165 - fillbetween-lorentzian1
166 - fillbetween-lorentzian2
167 comment: >
168 Plotter showing the model and the two coloured surfaces marking the components.
169
170 - kind: model
171 type: Polynomial
172 from_dataset: dummy
173 properties:
174 parameters:
175 coefficients: [0.1, 0.01]
176 result: baseline
177
178 - kind: processing
179 type: DatasetAlgebra
180 properties:
181 parameters:
182 kind: add
183 dataset: baseline
184 apply_to: model_data
185 result: model_data_with_baseline
186
187 - kind: processing
188 type: DatasetAlgebra
189 properties:
190 parameters:
191 kind: add
192 dataset: baseline
193 apply_to: lorentzian1
194 result: lorentzian1_with_baseline
195
196 - kind: processing
197 type: DatasetAlgebra
198 properties:
199 parameters:
200 kind: add
201 dataset: baseline
202 apply_to: lorentzian2
203 result: lorentzian2_with_baseline
204
205 - kind: multiplot
206 type: MultiPlotter1D
207 properties:
208 properties:
209 axes:
210 xlabel: "$position$ / a.u."
211 xlim: [0, 20]
212 drawings:
213 - color: black
214 linewidth: 1.5
215 - color: gray
216 linewidth: 1
217 parameters:
218 tight_layout: True
219 filename: plotting-annotation-fillbetween-baseline.pdf
220 apply_to:
221 - model_data_with_baseline
222 - baseline
223
224 - kind: plotannotation
225 type: FillBetween
226 properties:
227 parameters:
228 data:
229 - lorentzian1_with_baseline
230 second:
231 - baseline
232 properties:
233 facecolor: orange
234 alpha: 0.3
235 result: fillbetween-lorentzian1_with_baseline
236 comment: >
237 Coloured surfaces of the first Lorentzian.
238
239 - kind: plotannotation
240 type: FillBetween
241 properties:
242 parameters:
243 data:
244 - lorentzian2_with_baseline
245 second:
246 - baseline
247 properties:
248 facecolor: green
249 alpha: 0.3
250 result: fillbetween-lorentzian2_with_baseline
251 comment: >
252 Coloured surfaces of the second Lorentzian.
253
254 - kind: singleplot
255 type: SinglePlotter1D
256 properties:
257 properties:
258 axes:
259 xlabel: "$position$ / a.u."
260 xlim: [0, 20]
261 parameters:
262 tight_layout: True
263 filename: plotting-annotation-fillbetween-with-baseline.pdf
264 apply_to:
265 - model_data_with_baseline
266 annotations:
267 - fillbetween-lorentzian1_with_baseline
268 - fillbetween-lorentzian2_with_baseline
269 comment: >
270 Plotter showing the model and the two coloured surfaces
271 marking the components, but limited by the baseline.
Results
Examples for the figures created in the recipe are given below. While in the recipe, the output format has been set to PDF, for rendering them here they have been converted to PNG.
As this is a rather lengthy recipe demonstrating different scenarios, the individual cases are shown separately, each with the corresponding section of the recipe.
Setting the scene
The scenario: We have a curve comprising of two overlapping Lorentzians and want to highlight the individual components. Here, we just plot the data and the individual components.
49 parameters:
50 tight_layout: True
51 filename: plotting-annotation-marker-default.pdf
52 apply_to:
53 - model_data
54 result:
55 - plot-with-marker
56 comment: >
57 Plotter that gets annotated later
58
59 - kind: plotannotation
60 type: Marker
61 properties:
62 parameters:
63 xpositions: peaks
64 ypositions: 1.38
65 marker: "*"
66 plotter: plot-with-marker
67 comment: >
68 Star-shaped markers with default styling to highlight the peaks.
69
70 - kind: singleplot
71 type: SinglePlotter1D
72 properties:
The resulting figure is shown below:
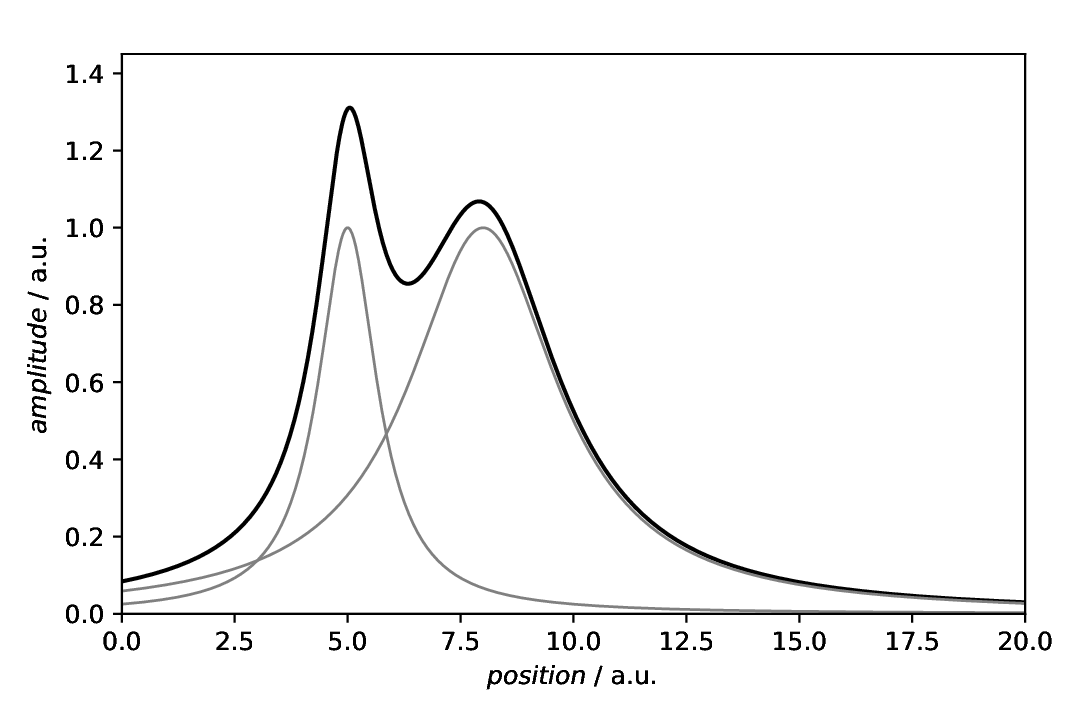
Fig. 35 Plot showing the composite model consisting of two Lorentzians together with the individual components. This is a typical scenario where one would like to add a shading below the curves of the individual components.
FillBetween with default settings
The scenario is as described above: We have a curve comprising of two overlapping Lorentzians and want to highlight the individual components. To fill the area below the curves of the individual components, we can use a aspecd.annotation.FillBetween annotation.
74 axes:
75 xlabel: "$position$ / a.u."
76 xlim: [0, 20]
77 ylim: [0, 1.45]
78 parameters:
79 tight_layout: True
80 filename: plotting-annotation-marker-styled.pdf
81 apply_to:
82 - model_data
83 result:
84 - plot-with-styled-marker
85 comment: >
86 Plotter that gets annotated later
87
88 - kind: plotannotation
89 type: Marker
90 properties:
91 parameters:
92 xpositions: peaks
93 ypositions: 1.38
94 marker: "h"
95 properties:
96 edgecolor: red
97 edgewidth: 2
98 facecolor: blue
99 facecoloralt: green
100 size: 16
101 fillstyle: top
102 alpha: 0.8
The resulting figure is shown below:
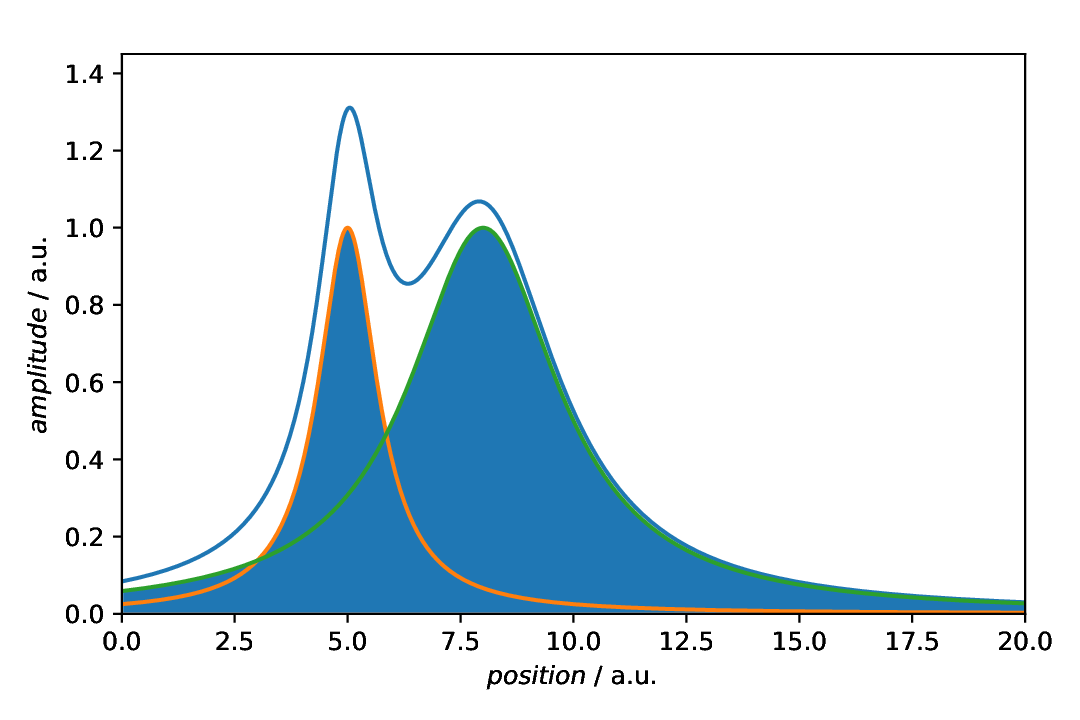
Fig. 36 Plot with shading of the individual components as annotation. Note that the individual components are plotted explicitly as well, using a aspecd.plotting.MultiPlotter1D.
The default settings are most probably not exactly what we usually want to have. First of all, individual components should often been marked with different colours, and adding a bit of transparency may help as well. Off we go.
Individual annotations with styling
In the example above, we have shown the default styling of the aspecd.annotation.FillBetween annotation. As this is not quite what we usually would like to see, here, we apply some styling. For details of the parameters you can set, have a look at aspecd.plotting.PatchProperties.
104 comment: >
105 Styled markers demonstrating some of the styling possibilities.
106
107 - kind: singleanalysis
108 type: PeakFinding
109 properties:
110 parameters:
111 return_intensities: True
112 apply_to: model_data
113 result: peaks_xy
114
115 - kind: singleplot
116 type: SinglePlotter1D
117 properties:
118 properties:
119 axes:
120 xlabel: "$position$ / a.u."
121 xlim: [0, 20]
122 ylim: [0, 1.45]
123 parameters:
124 tight_layout: True
125 filename: plotting-annotation-marker-peaks.pdf
126 apply_to:
127 - model_data
128 result:
129 - plot-with-marker-peaks
130 comment: >
131 Plotter that gets annotated later
132
133 - kind: plotannotation
134 type: Marker
135 properties:
136 parameters:
137 positions: peaks_xy
138 marker: "*"
139 plotter: plot-with-marker-peaks
140 comment: >
141 Star-shaped markers with default styling placed on peaks.
142
143 - kind: singleplot
144 type: SinglePlotter1D
145 properties:
146 properties:
147 axes:
148 xlabel: "$position$ / a.u."
149 xlim: [0, 20]
The resulting figure is shown below:
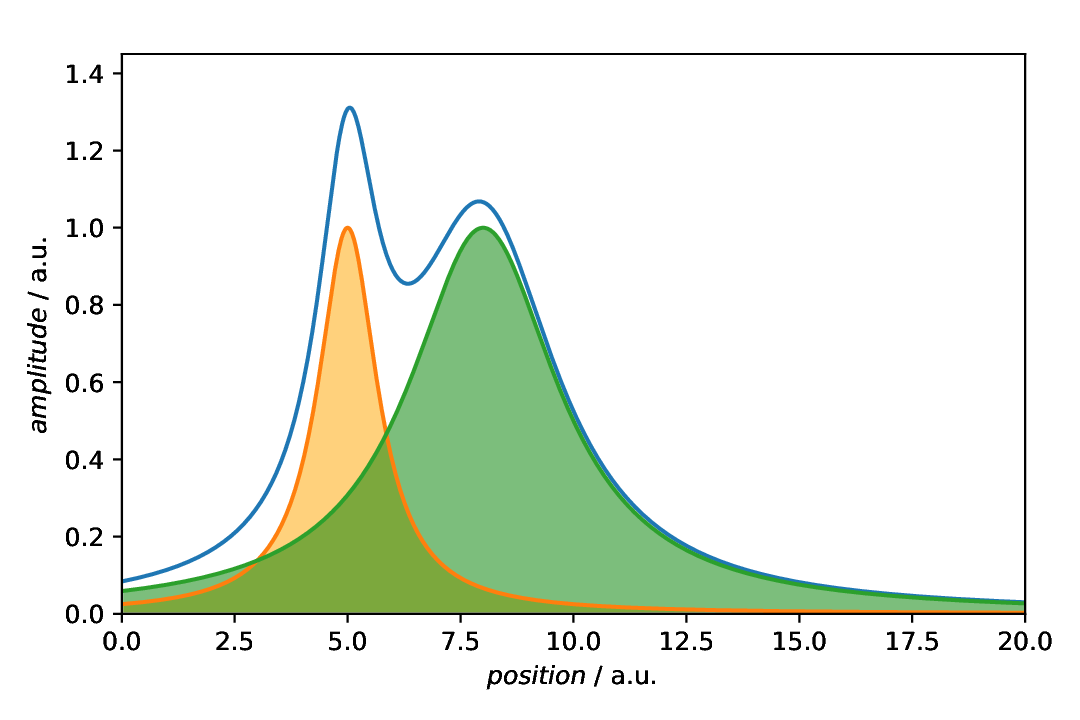
Fig. 37 Plot with both components styled individually. Here, we applied a colour (note the difference between facecolor and edgecolor for this type of annotation) and some transparency.
Individual annotations in a SinglePlotter
Quite similar to the situation above, we need not use a aspecd.plotting.MultiPlotter to show the individual components, as the annotation would be sufficient. Note that we stored the annotations for the individual lines declared above using the result key, so that we can reuse them here.
151 parameters:
152 tight_layout: True
153 filename: plotting-annotation-marker-peaks-yoffset.pdf
154 apply_to:
155 - model_data
156 result:
157 - plot-with-marker-peaks-yoffset
158 comment: >
159 Plotter that gets annotated later
160
161 - kind: plotannotation
162 type: Marker
163 properties:
164 parameters:
165 positions: peaks_xy
166 yoffset: 0.05
167 marker: "*"
168 plotter: plot-with-marker-peaks-yoffset
The resulting figure is shown below:
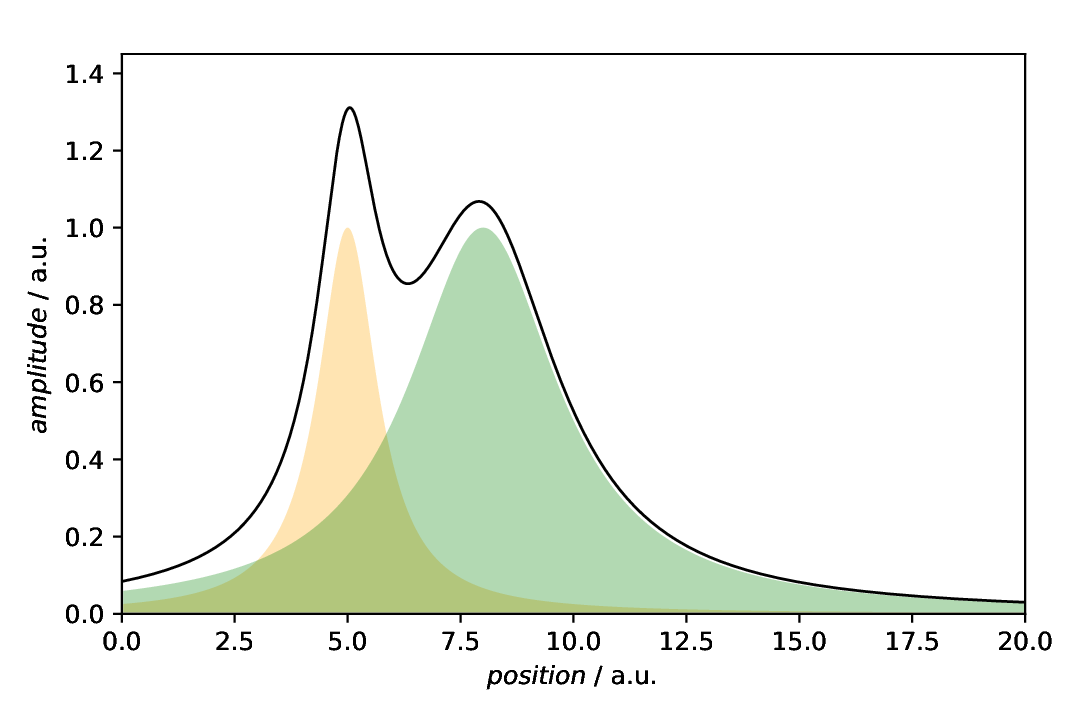
Fig. 38 Plot showing only the line of the composite model, annotated with the shaded areas of the indivual components.
Data with baseline
By default, and if not providing a second line, the patch extends from zero to the data points given by the actual data of the curve. While you could set a scalar value different from zero, more often you will encounter baselines, at least in spectroscopy and in real life. Here, we first (re)create the same data as above, but this time with a baseline added. To add the baseline to both, the model data and the individual components, we use the aspecd.processing.DatasetAlgebra processing step. Afterwards, the data with baseline are plotted together with the baseline.
170 Star-shaped markers with default styling placed on peaks, vertically offset.
171
172 - kind: singleplot
173 type: SinglePlotter1D
174 properties:
175 properties:
176 axes:
177 xlabel: "$position$ / a.u."
178 xlim: [0, 20]
179 ylim: [0, 1.45]
180 parameters:
181 tight_layout: True
182 filename: plotting-annotation-marker-by-name.pdf
183 apply_to:
184 - model_data
185 result:
186 - plot-with-marker-by-name
187 comment: >
188 Plotter that gets annotated later
189
190 - kind: plotannotation
191 type: Marker
192 properties:
193 parameters:
194 positions: peaks_xy
195 yoffset: 0.005
196 marker: "caretdown"
197 properties:
198 edgewidth: 0
199 plotter: plot-with-marker-by-name
200 comment: >
201 Markers with default styling identified by their name.
202
203 - kind: singleplot
204 type: SinglePlotter1D
205 properties:
206 properties:
207 axes:
208 xlabel: "$position$ / a.u."
209 xlim: [0, 20]
210 ylim: [0, 1.45]
211 parameters:
212 tight_layout: True
213 filename: plotting-annotation-marker-mathtext.pdf
214 apply_to:
215 - model_data
216 result:
217 - plot-with-marker-mathtext
218 comment: >
219 Plotter that gets annotated later
220
221 - kind: plotannotation
222 type: Marker
The resulting figure is shown below:
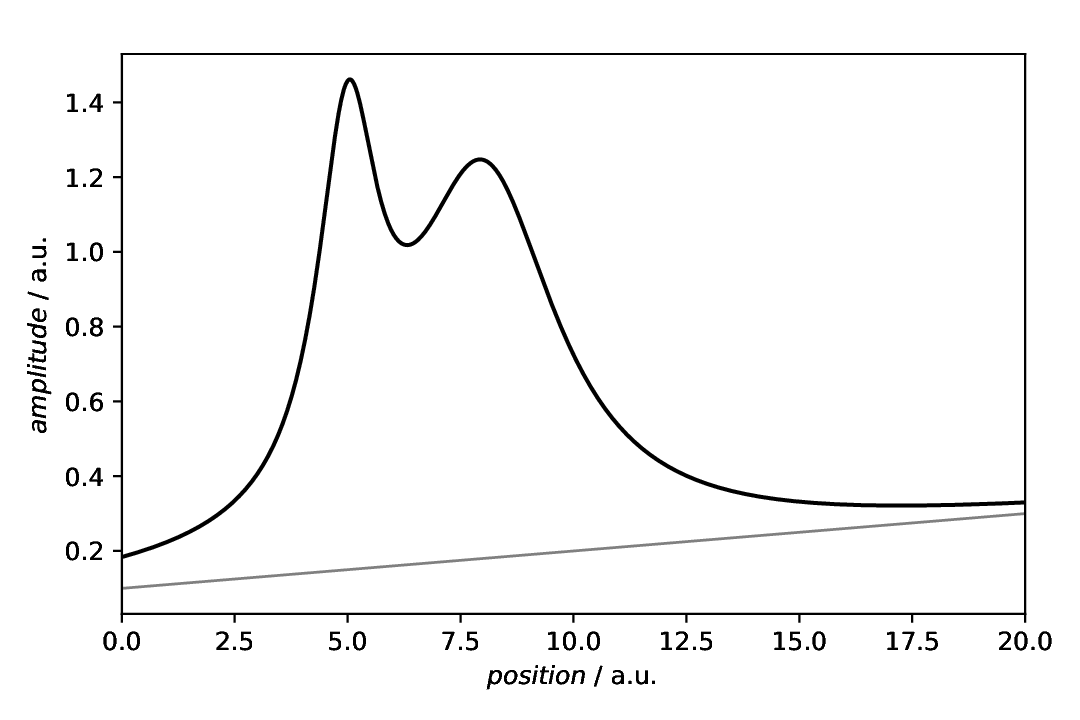
Fig. 39 Data with a baseline added, together with the baseline. A bit of styling is applied here, to show that the baseline is just an addition, not the actual data of interest.
Filling between two curves
Having data with a baseline, this is finally a sensible real-world situation where you may want to provide a second dataset as boundary for the patch. Of course, other typical scenarios are marking the confidence interval of a fit, where you have the lower and upper boundaries as dataset.
224 parameters:
225 positions: peaks_xy
226 yoffset: 0.05
227 marker: $\mathcal{A}$
228 properties:
229 size: 14
230 edgewidth: 0
231 facecolor: orange
232 plotter: plot-with-marker-mathtext
233 comment: >
234 Markers with default styling using MathText (no LaTeX install needed).
235
236 Note that in this case, you cannot have question marks surrounding the
237 marker string, as otherwise, YAML is unhappy.
238
239 - kind: singleplot
240 type: SinglePlotter1D
241 properties:
242 properties:
243 axes:
244 xlabel: "$position$ / a.u."
245 xlim: [0, 20]
246 ylim: [0, 1.45]
247 parameters:
248 tight_layout: True
249 filename: plotting-annotation-marker-unicode.pdf
250 apply_to:
251 - model_data
252 result:
253 - plot-with-marker-unicode
254 comment: >
255 Plotter that gets annotated later
256
257 - kind: plotannotation
258 type: Marker
259 properties:
260 parameters:
261 positions: peaks_xy
262 yoffset: 0.06
263 marker: "$\u266B$"
264 properties:
265 size: 14
266 edgewidth: 0
267 facecolor: blue
268 plotter: plot-with-marker-unicode
269 comment: >
270 Markers with default styling using Unicode (there is music in the peaks).
271
272 Note that in this case, you need to have question marks surrounding the
The resulting figure is shown below:
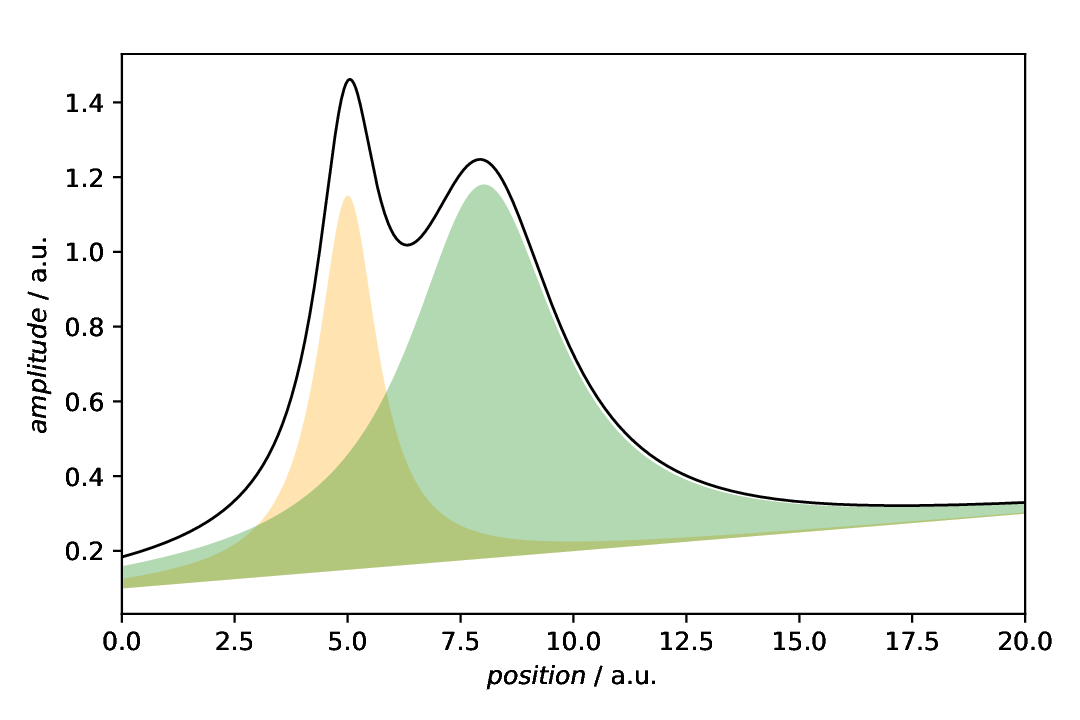
Fig. 40 Data with baseline together with the individual components, but respecting the baseline as (in this case lower) boundary of the fill patch.
Comments
As usual, a model dataset is created at the beginning, to have something to show. Here, a CompositeModel comprising of two Lorentizans is used to get two peaks that can be labelled.
Furthermore, the individual Lorentzians are created as well.
For simplicity, generic plotters are used, to focus on the annotations.
The sequence of defining plot and annotation(s) does not matter. You only need to provide the
resultkey with a unique name for whichever task you define first, to refer to it in the later task(s).